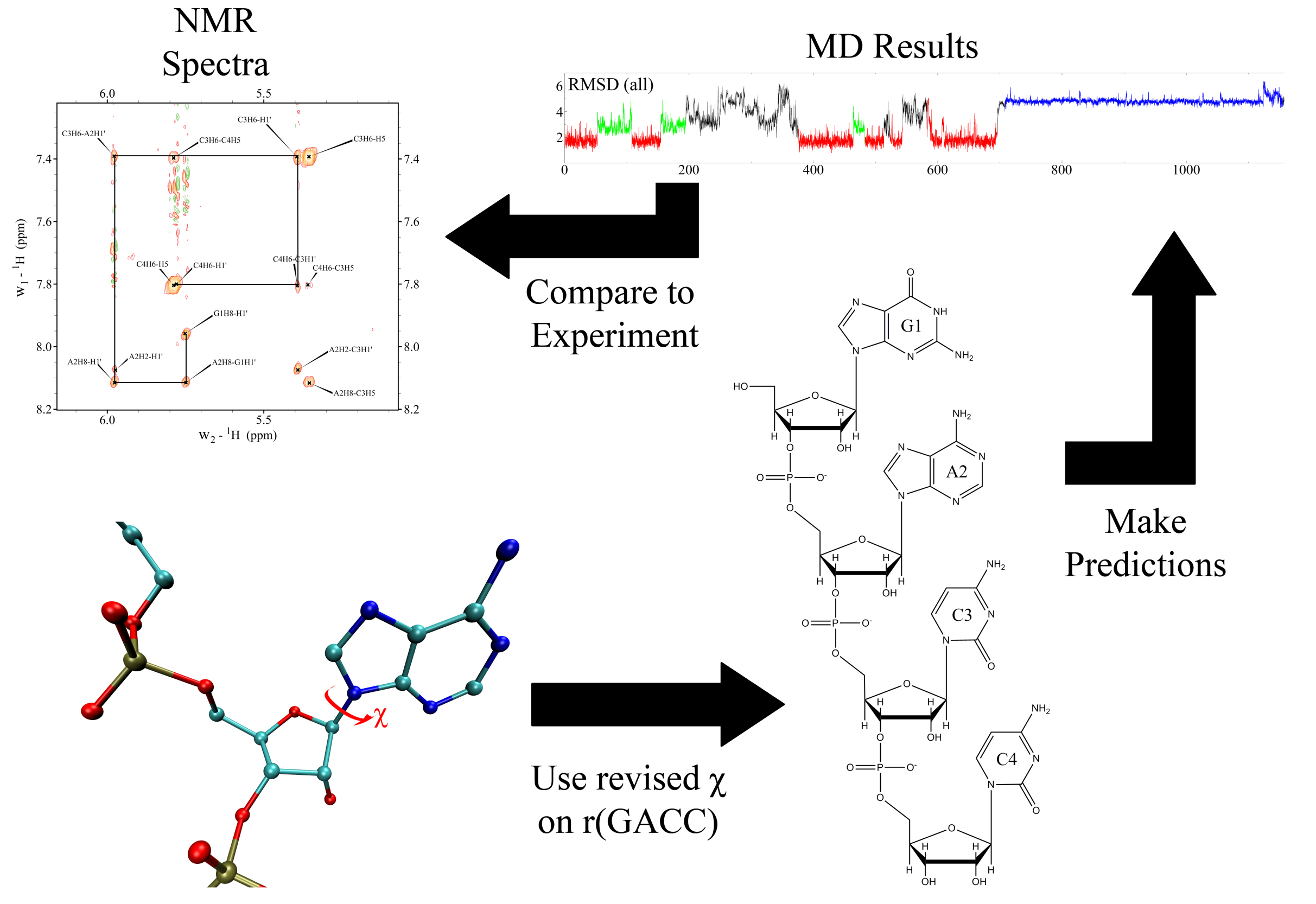
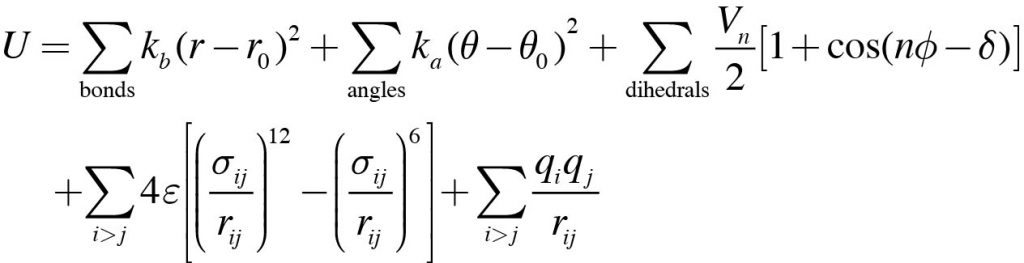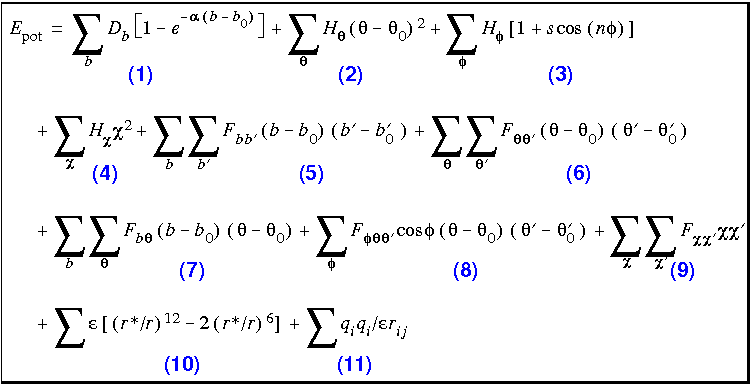CHARMM Force-Field Parameters for Morphine, Heroin, and Oliceridine, and Conformational Dynamics of Opioid Drugs | Journal of Chemical Information and Modeling

PMFF: Development of a Physics-Based Molecular Force Field for Protein Simulation and Ligand Docking | The Journal of Physical Chemistry B

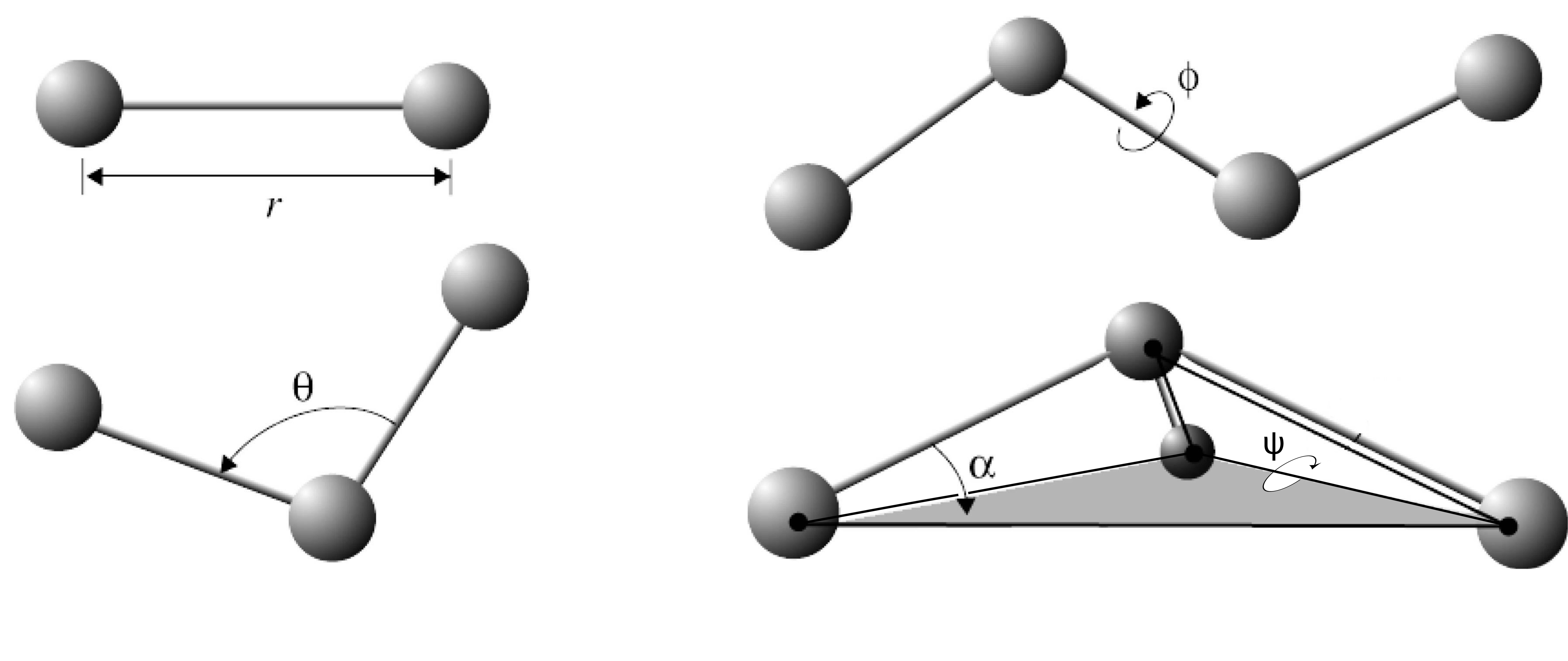
1: Schematic illustration of the bonded terms in the CHARMM force field... | Download Scientific Diagram
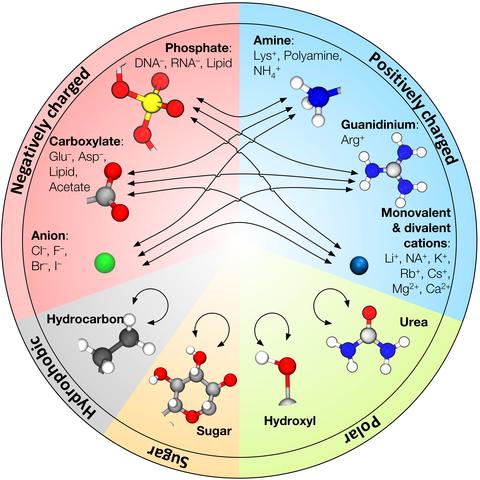
CUFIX: Non-bonded Fix (NBFIX) parameters for the CHARMM and AMBER force fields | The Aksimentiev Group
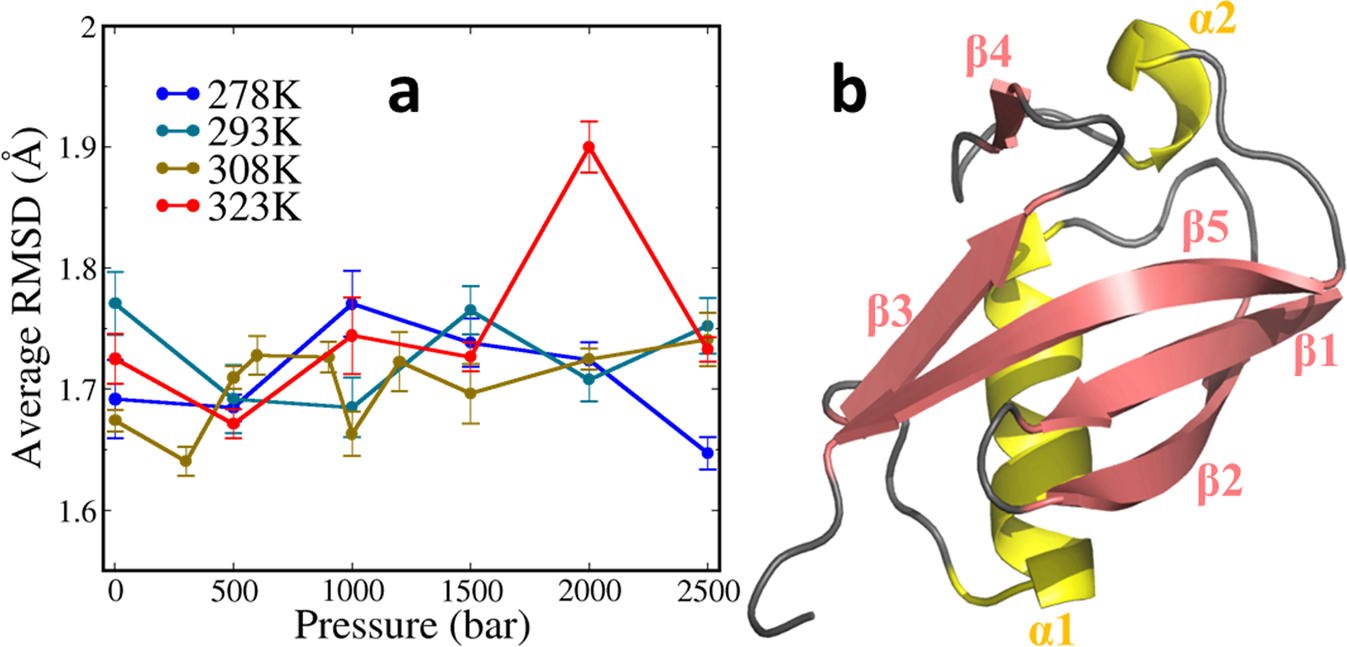
Validating the CHARMM36m protein force field with LJ-PME reveals altered hydrogen bonding dynamics under elevated pressures | Communications Chemistry

An Empirical Polarizable Force Field Based on the Classical Drude Oscillator Model: Development History and Recent Applications. - Abstract - Europe PMC

CHARMM force field and molecular dynamics simulations of protonated polyethylenimine - Beu - 2017 - Journal of Computational Chemistry - Wiley Online Library
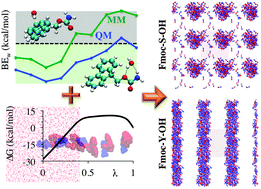
CHARMM force field parameterization protocol for self-assembling peptide amphiphiles: the Fmoc moiety - Physical Chemistry Chemical Physics (RSC Publishing)

Table 1 from CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field | Semantic Scholar

Webinar: CHARMM Force Field Development History, Features, and Implementation in GROMACS (2021-11-23) – BioExcel – Centre of Excellence for Computation Biomolecular Research

BioExcel Webinar #58: CHARMM Force Field Development History, Features and Implementation in GROMACS - YouTube
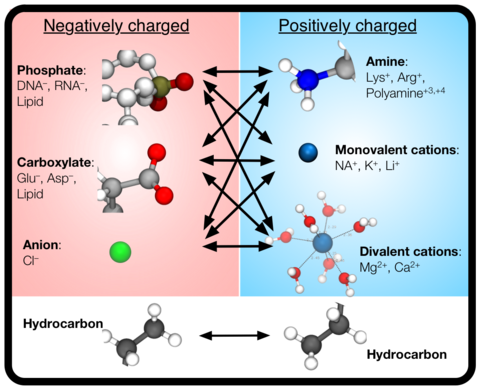
CUFIX: Non-bonded Fix (NBFIX) parameters for the CHARMM and AMBER force fields | The Aksimentiev Group

The determination of CHARMM force field parameters for the Mg2+ containing HIV-1 integrase - ScienceDirect
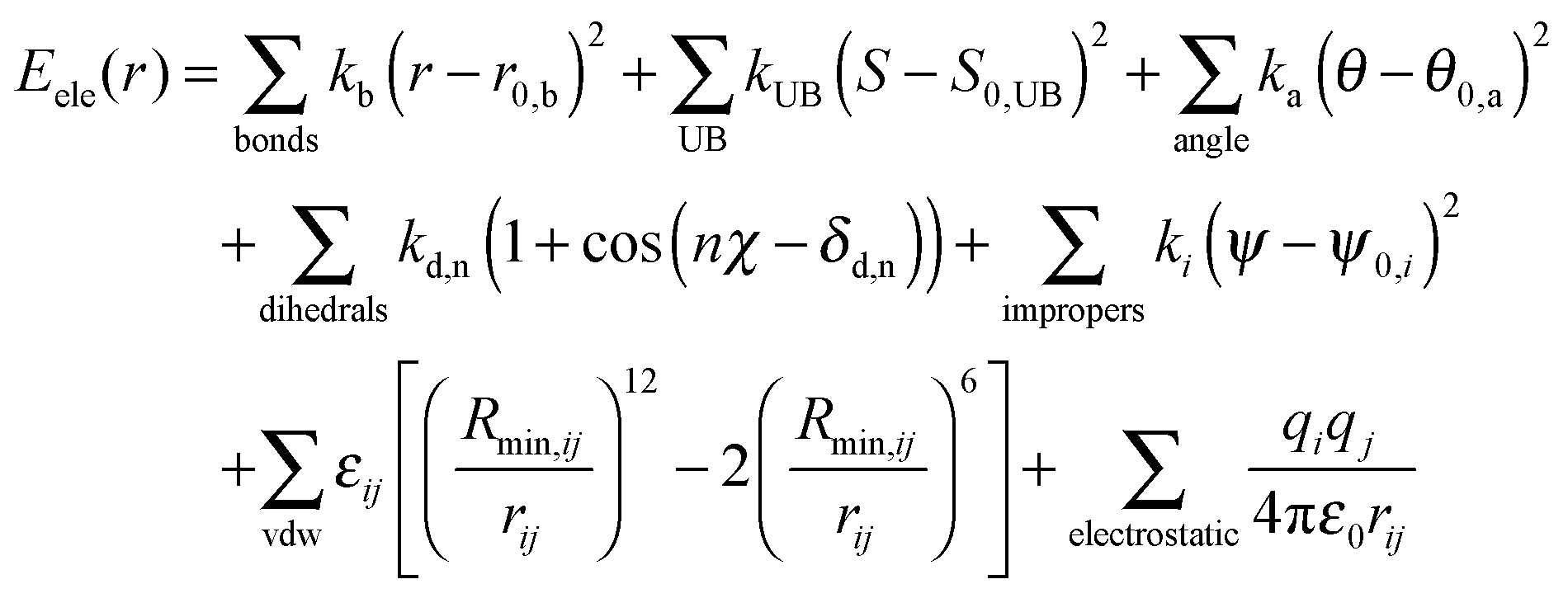
CHARMM force field parameterization protocol for self-assembling peptide amphiphiles: the Fmoc moiety - Physical Chemistry Chemical Physics (RSC Publishing) DOI:10.1039/C5CP06770G

![PDF] Development of the CHARMM Force Field for Lipids. | Semantic Scholar PDF] Development of the CHARMM Force Field for Lipids. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/d626430abc91542bde9a9744cada295af20b5c71/3-Table1-1.png)